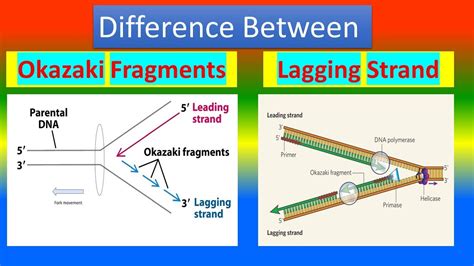
Understanding DNA Replication: The Importance of Okazaki Fragments

DNA replication is a complex process that involves the duplication of genetic material, ensuring the continuity of genetic information from one generation to the next. The replication process is semi-conservative, meaning that each new DNA molecule contains one old strand (the template strand) and one newly synthesized strand. During this process, short, discontinuous DNA segments called Okazaki fragments are formed on the lagging strand. The lagging strand is the strand that runs in the opposite direction to the leading strand, and its replication is more complicated due to the antiparallel nature of DNA.
The formation of Okazaki fragments is a crucial aspect of DNA replication, as it allows for the synthesis of the lagging strand in short, manageable segments. This process is mediated by enzymes such as DNA polymerase and primase, which work together to synthesize new DNA strands. In this article, we will explore the five ways Okazaki fragments form on the lagging strand, highlighting the key players and mechanisms involved.
1. Initiation of Okazaki Fragment Synthesis

The synthesis of Okazaki fragments begins with the unwinding of the double helix structure of DNA at the replication fork. This is mediated by the enzyme helicase, which breaks the hydrogen bonds between the two strands. As the leading strand is synthesized continuously, the lagging strand is replicated in short, discontinuous segments. The initiation of Okazaki fragment synthesis requires the presence of a primer, which is a short RNA molecule that provides a starting point for DNA synthesis.
Primase, an enzyme responsible for adding RNA primers to the template strand, plays a crucial role in the initiation of Okazaki fragment synthesis. The primer is complementary to the template strand and provides a free 3' hydroxyl group, which is necessary for DNA polymerase to extend the new DNA strand.
Key Players Involved in Okazaki Fragment Initiation
- Helicase: unwinds the double helix structure of DNA
- Primase: adds RNA primers to the template strand
- DNA polymerase: extends the new DNA strand using the primer as a starting point
2. Elongation of Okazaki Fragments

Once the primer is in place, DNA polymerase can begin to elongate the Okazaki fragment. This process involves the addition of nucleotides to the growing DNA strand, using the template strand as a guide. DNA polymerase reads the template strand and matches the incoming nucleotides to the base pairing rules (A-T and G-C).
The elongation of Okazaki fragments is a rapid process, with DNA polymerase able to add nucleotides at a rate of up to 1,000 per minute. However, this process is not error-free, and mistakes can occur during the synthesis of new DNA strands. Fortunately, DNA polymerase has proofreading capabilities, allowing it to correct errors as they occur.
Key Features of Okazaki Fragment Elongation
- DNA polymerase reads the template strand and adds nucleotides to the growing DNA strand
- Base pairing rules (A-T and G-C) guide the addition of nucleotides
- Proofreading capabilities allow DNA polymerase to correct errors during synthesis
3. Ligation of Okazaki Fragments

As the Okazaki fragments are synthesized, they must be ligated together to form a continuous DNA strand. This process is mediated by the enzyme DNA ligase, which seals the gaps between the Okazaki fragments.
DNA ligase forms a phosphodiester bond between the 3' end of one Okazaki fragment and the 5' end of the next, effectively linking the fragments together. This process is essential for maintaining the integrity of the DNA molecule and ensuring that the genetic information is accurately replicated.
Key Features of Okazaki Fragment Ligation
- DNA ligase seals the gaps between Okazaki fragments
- Phosphodiester bond forms between the 3' end of one Okazaki fragment and the 5' end of the next
4. RNA Primer Removal

After the Okazaki fragments have been ligated together, the RNA primers must be removed. This process is mediated by the enzyme RNase H, which specifically degrades RNA-DNA hybrids.
RNase H cleaves the RNA primer at the junction between the RNA and DNA, creating a free 3' hydroxyl group. This allows DNA polymerase to fill in the gap with DNA nucleotides, effectively replacing the RNA primer.
Key Features of RNA Primer Removal
- RNase H degrades RNA-DNA hybrids
- Cleavage occurs at the junction between the RNA and DNA
- DNA polymerase fills in the gap with DNA nucleotides
5. Completion of Lagging Strand Synthesis

The final stage of Okazaki fragment synthesis involves the completion of the lagging strand. This process requires the ligation of the remaining Okazaki fragments and the removal of any remaining RNA primers.
Once the lagging strand is complete, the replication process is finished, and the new DNA molecule is formed. The new molecule contains one old strand (the template strand) and one newly synthesized strand, ensuring that the genetic information is accurately replicated.
Key Features of Lagging Strand Synthesis Completion
- Ligation of remaining Okazaki fragments
- Removal of remaining RNA primers
- Completion of the lagging strand
We hope this article has provided a comprehensive overview of the five ways Okazaki fragments form on the lagging strand. The synthesis of Okazaki fragments is a crucial aspect of DNA replication, and understanding the mechanisms involved can provide valuable insights into the complex process of genetic information replication. Share your thoughts and comments below!
What is the role of Okazaki fragments in DNA replication?
+Okazaki fragments are short, discontinuous DNA segments synthesized on the lagging strand during DNA replication. They allow for the synthesis of the lagging strand in short, manageable segments.
What is the function of DNA ligase in Okazaki fragment synthesis?
+DNA ligase seals the gaps between Okazaki fragments, forming a phosphodiester bond between the 3' end of one Okazaki fragment and the 5' end of the next.
What is the purpose of RNase H in RNA primer removal?
+RNase H specifically degrades RNA-DNA hybrids, cleaving the RNA primer at the junction between the RNA and DNA.