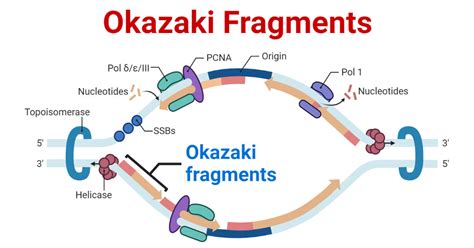
The process of DNA replication is a complex and highly regulated mechanism that ensures the accurate duplication of genetic material. One crucial aspect of this process is the formation of Okazaki fragments, which are short, discontinuous DNA segments synthesized on the lagging strand during replication. Understanding the reasons behind Okazaki fragment formation is essential to grasping the intricacies of DNA replication. Here, we will delve into the five primary reasons why Okazaki fragments form.
Reason 1: Unidirectional DNA Synthesis

DNA synthesis is a unidirectional process, meaning that it occurs in one direction, from the 5' end to the 3' end of the DNA strand. This directionality is due to the enzyme DNA polymerase, which can only add nucleotides to the 3' end of a growing DNA strand. As a result, the leading strand is synthesized continuously, while the lagging strand is synthesized in short, discontinuous segments – the Okazaki fragments.
Leading Strand vs. Lagging Strand
The leading strand is the template strand that runs in the 3' to 5' direction, allowing DNA polymerase to synthesize the new DNA strand continuously. In contrast, the lagging strand runs in the opposite direction, from 5' to 3'. To accommodate this directionality, the lagging strand is synthesized in short segments, which are later joined together to form the complete strand.
Reason 2: RNA Primers and Initiation of DNA Synthesis

The initiation of DNA synthesis on the lagging strand requires the presence of RNA primers. These short RNA molecules provide a starting point for DNA polymerase to begin synthesizing the new DNA strand. However, RNA primers are only temporary and are eventually removed and replaced with DNA. The replacement process occurs in short segments, resulting in the formation of Okazaki fragments.
RNA Primer Removal and Replacement
Once DNA synthesis is initiated on the lagging strand, the RNA primer is removed, and the resulting gap is filled with DNA. This process is repeated multiple times, resulting in the formation of multiple Okazaki fragments. The fragments are then joined together through a process called ligation, which involves the enzyme DNA ligase.
Reason 3: Limited Processivity of DNA Polymerase

DNA polymerase, the enzyme responsible for synthesizing new DNA strands, has limited processivity. This means that it can only synthesize a certain length of DNA before releasing the template strand and re-initiating synthesis at a new location. On the lagging strand, this limited processivity results in the formation of Okazaki fragments.
Processivity and the Length of Okazaki Fragments
The length of Okazaki fragments is influenced by the processivity of DNA polymerase. In prokaryotic cells, Okazaki fragments are typically around 1-2 kilobase pairs (kbp) in length, while in eukaryotic cells, they are shorter, ranging from 100-200 base pairs (bp). The limited processivity of DNA polymerase ensures that the lagging strand is synthesized in short, manageable segments.
Reason 4: Coordination with the Replication Fork

The replication fork is the region where the DNA double helix is unwound, and the leading and lagging strands are separated. The formation of Okazaki fragments on the lagging strand must be coordinated with the movement of the replication fork. This ensures that the lagging strand is synthesized at the correct rate and that the replication process proceeds smoothly.
Helicase and the Replication Fork
The enzyme helicase is responsible for unwinding the DNA double helix at the replication fork. As the replication fork moves forward, the lagging strand is synthesized in short segments, allowing the leading strand to be continuously synthesized. The coordination between helicase and the synthesis of Okazaki fragments ensures that the replication process is efficient and accurate.
Reason 5: Ensuring Accuracy and Fidelity

The formation of Okazaki fragments ensures that the replication process is accurate and faithful. By synthesizing the lagging strand in short segments, errors can be corrected more efficiently, and the risk of mutations is reduced. This is particularly important in regions with high GC content, where the formation of Okazaki fragments allows for more accurate synthesis.
Error Correction and Proofreading
The synthesis of Okazaki fragments allows for more efficient error correction and proofreading. As each fragment is synthesized, the enzyme DNA polymerase can correct errors and ensure that the resulting DNA strand is accurate. This process is essential for maintaining the integrity of the genome and preventing mutations.
In conclusion, the formation of Okazaki fragments is a critical aspect of DNA replication, ensuring that the lagging strand is synthesized accurately and efficiently. The five reasons outlined above highlight the importance of Okazaki fragments in maintaining the integrity of the genome and ensuring the fidelity of the replication process.
We encourage you to share your thoughts and insights on the importance of Okazaki fragments in the comments section below. What do you think is the most significant reason for the formation of Okazaki fragments? Share your ideas and let's start a conversation!
What is the primary function of Okazaki fragments in DNA replication?
+The primary function of Okazaki fragments is to allow for the synthesis of the lagging strand in short, discontinuous segments, ensuring accurate and efficient DNA replication.
How do RNA primers contribute to the formation of Okazaki fragments?
+RNA primers provide a starting point for DNA polymerase to initiate DNA synthesis on the lagging strand. The primers are eventually removed and replaced with DNA, resulting in the formation of Okazaki fragments.
What is the role of helicase in the formation of Okazaki fragments?
+Helicase unwinds the DNA double helix at the replication fork, allowing the lagging strand to be synthesized in short segments, which are then joined together to form Okazaki fragments.